lipoyl domain of the dihydrolipoyl acetyltransferase subunit from Escherichia coli (E2lip3( )): pulling from C-ter to Residue 41 (Lys)
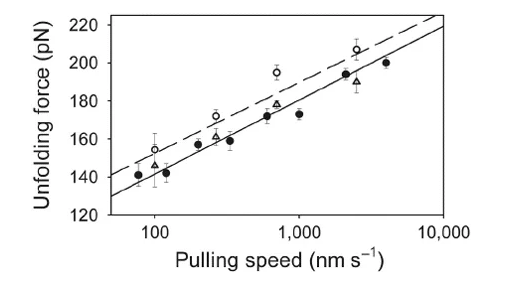
Figure: The mean unfolding force of triplicate data sets ( s.e.m.) at 100, 266, 700 and 2,500 nm s-1 for I27 (empty circle) and E2lip3 (triangle) in (I27)4E2lip3(+). The speed dependence for (I27)5* (ref. 7) is shown for comparison (filled circle). Continuous and discontinuous lines show the speed dependence of mechanical unfolding predicted for E2lip3(+) and I27 in (I27)4E2lip3(+), that is, four domains of I27 and one of E2lip3(+) using a Monte Carlo simulation. Both the mean unfolding force and the speed dependence of the unfolding force for E2lip3(+) are experimentally indistinguishable from those obtained for (I27)5* , even though E2lip3(+) would be expected to unfold at a lower force than I27 if each domain was concatamerized into homopolymers of identical length. This results from the number and length of previously unfolded domains that markedly affect the observed unfolding force
Reference: David J Brockwell, Emanuele Paci, Rebecca C Zinober, Godfrey S Beddard, Peter D Olmsted, D Alastair Smith, Richard N Perham, Sheena E Redford et al. 2003 (https://doi.org/10.1038/nsb968 )
Reference: David J Brockwell, Emanuele Paci, Rebecca C Zinober, Godfrey S Beddard, Peter D Olmsted, D Alastair Smith, Richard N Perham, Sheena E Redford et al. 2003 (https://doi.org/10.1038/nsb968 )