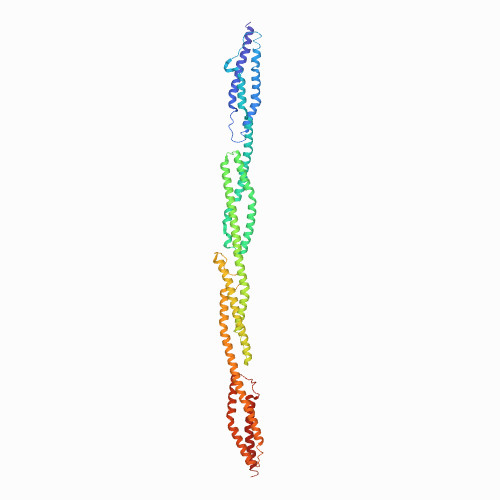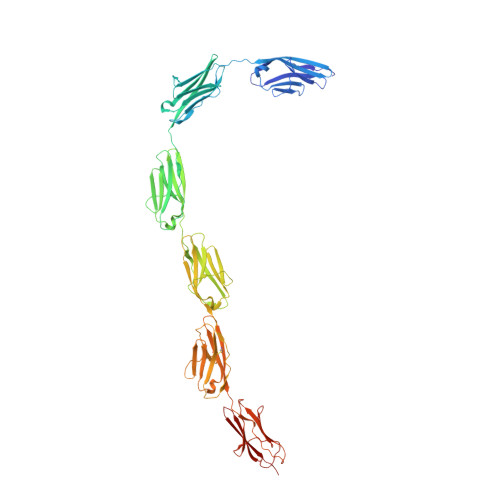| Actinomyces fimbrial adhesin FimA | 3QDH | Mainly Beta | 500 |
| 0.00e+00 | 100
| N/A | N/A | 14.2 | N/A | 20 | Danature pulse, buffer: calcium (10 mM) to EDTA (1 mM) | 272 | 200-472 |  | Actinomyces naeslundii | No | CELL ADHESION | AFM | constant force | DOI | 2011-08-24 | Devarajan, B., Krishnan, V., Narayana, S.V.L. | Daniel J. Echelman, Jorge Alegre-Cebollada, Carmen L. Badilla, and Julio M. Fernández - 2016 | | N_U[14] |  | 0.58 | |
| bovine carbonic anhydrase B (CAB) (it is also called bovine carbonic dehydratase II) under native conditions (with knot) | 1V9E | All beta proteins | 1700 | 200
| 1.50e+02 |
| N/A | N/A | 13 | N/A | N/A | a 50 mM Tris SO4^2+ buffer at pH 7.3 | 261 | 1-261 |  | Bos taurus | No | LYASE | AFM | constant velocity | DOI | 2004-02-10 | Saito, R., Sato, T., Ikai, A., Tanaka, N. | T Wang, H Arakawa, A Ikai - 2001 | | N_U[13] |  | | |
| bovine carbonic anhydrase B (CAB) (it is also called
bovine carbonic dehydratase II) without knot (knotless) | 1V9E | All beta proteins | 200 | 1
| 1.50e+02 |
| N/A | N/A | 110 | N/A | N/A | a 50 mM Tris SO4^2+ buffer at pH 7.3 | 261 | 1-261 |  | Bos taurus | No | LYASE | AFM | constant velocity | DOI | 2004-02-10 | Saito, R., Sato, T., Ikai, A., Tanaka, N. | T Wang, H Arakawa, A Ikai - 2001 | | N_U[110] |  | | |
| Phytochrome carrying its covalently bound tetrapyrrole chromophore (DrBphP CBD) | 2O9C | Alpha and beta proteins (a+b) | 73 | 1
| 1.00e+07 |
| N/A | N/A | 104 | N/A | N/A | ff03 force field at fixed pressure (1 bar) and temperature (300 K), NPT ensemble, periodic boundary, constant velocity | | 4-322 |  | Deinococcus radiodurans | Yes | TRANSFERASE | all atom SMD simulation | constant velocity | DOI | 2007-03-06 | Wagner, J.R., Brunzelle, J.S., Vierstra, R.D., Forest, K.T. | Thomas Bornschlogl, David M. Anstrom, Elisabeth Mey, Joachim Dzubiella, Matthias Rief, Katrina T. Forest - 2008 | | N_U[104] |  | | |
| Phytochrome (apo-DrCBD) | 2O9C | Alpha and beta proteins (a+b) | 47 | 1
| 1.00e+03 |
| N/A | N/A | 98.1 | N/A | N/A | 0.5 mg/mL in phosphate-buffered saline | | 4-322 |  | Deinococcus radiodurans | Yes | TRANSFERASE | AFM | constant velocity | DOI | 2007-03-06 | Wagner, J.R., Brunzelle, J.S., Vierstra, R.D., Forest, K.T. | Thomas Bornschlogl, David M. Anstrom, Elisabeth Mey, Joachim Dzubiella, Matthias Rief, Katrina T. Forest - 2008 | | N_U[98] |  | | |
| R16 repeat of Spectrin | 1AJ3 | All alpha proteins | 70 | 10
| 3000 |
| N/A | N/A | 31 | N/A | 20 | buffer 50 mM Tris HCl pH 8.0, 50 mM NaCl | 106 | 1-106 |  | Gallus gallus | Yes | CYTOSKELETON | AFM | constant velocity | DOI | 1997-07-07 | Pascual, J., Pfuhl, M., Walther, D., Saraste, M., Nilges, M. | P.-F Lenne, A.J Raae, S.M Altmann, M Saraste, J.K.H Horber - 2000 | N/A | N_I[15.5]_U[31] |  | | |
| Alpha 21 domain of Human Spectrin | 1S35 | All alpha proteins | 27 | 1
| 1.00e+03 |
| N/A | N/A | 41 | 1.0E-5 | 23 | PBS buffer | 111 | 1063-1264 |  | Homo sapiens | Yes | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2004-04-20 | Kusunoki, H., MacDonald, R.I., Mondragon, A. | Richard Law, Philippe Carl, Sandy Harper, Paul Dalhaimer, David W. Speicher, Dennis E. Discher - 2003 | | N_U[41] |  | | |
| Alpha-actin | ACTININ | 1HCI | All alpha proteins | 38 |
| 3.00e+02 |
| N/A | N/A | | N/A | 20 | PBS | 746 | 1-746 |  | Homo sapiens | No | TRIPLE-HELIX COILED COIL | AFM | constant velocity | DOI | 2001-06-27 | Ylanne, J., Scheffzek, K., Young, P., Saraste, M. | Richard Law, Sandy Harper, David W Speicher, Dennis E Discher - 2004 | | |  | | |
| wild-type Beta2-microglobulin (B2m) | 1LDS | All beta proteins | 146 | 27
| 2.18e+03 |
| N/A | N/A | 14 | N/A | 20 | PBS 10 mM | 98 | 1-99 |  | Homo sapiens | No | IMMUNE SYSTEM | AFM | constant velocity | DOI | 1998-04-08 | Trinh, C.H., Smith, D.P., Kalverda, A.P., Phillips, S.E.V., Radford, S.E. | B. Sorce, S. Sabella, M. Sandal, B. Samori, A. Santino, R. Cingolani, R. Rinaldi, P. P. Pompa - 2009 | NA | N_U[14] |  | | |
| Mutant of Beta2-microglobulin (B2m): Deletion of 6 N-ter amino acids | 1LDS | All beta proteins | 80 | 17
| 2.18e+03 |
| N/A | N/A | 11.5 | N/A | 20 | PBS 10 mM | 92 | 1-93 |  | Homo sapiens | Yes | IMMUNE SYSTEM | AFM | constant velocity | DOI | 1998-04-08 | Trinh, C.H., Smith, D.P., Kalverda, A.P., Phillips, S.E.V., Radford, S.E. | B. Sorce, S. Sabella, M. Sandal, B. Samori, A. Santino, R. Cingolani, R. Rinaldi, P. P. Pompa - 2009 | NA | N_U[11.5] |  | | |
| GFP polyprotein linked by cysteine on 3-132| 1EMB: GREEN FLUORESCENT PROTEIN (GFP) FROM AEQUOREA VICTORIA, GLN 80 REPLACED WITH ARG | 1EMB | Alpha and beta proteins (a+b) | 350 | 10
| 3.60e+03 |
| N/A | N/A | 41.6 | N/A | N/A | PBS buffer (pH 7.4) at high protein concentrations of ~5 mg/ml (~0.2 mM) - Different pulling directions: see more in name column | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| Protein ribonuclease HI Q4C/V155C of E.coli | 2YV0 | Alpha and beta proteins (a/b) | 18 | 1
| 300 | 53
See more | N/A | 5.1 | 41 | 0.00485 | 20 | 250 mM NaCl, 10 mM Tris, pH 7.0, and 1 mM EDTA | 155 | 1-155 |  | Escherichia coli | Yes | HYDROLASE | Optical Tweezers (OT) | constant force | DOI | 2008-03-18 | Haruki, M., Motegi, T., Tadokoro, T., Koga, Y., Takano, K., Kanaya, S. | Ciro Cecconi, Elizabeth A Shank, Carlos Bustamante, Susan Marqusee - 2005 | | N_I_I_U[41] |  | | |
| full titin | uniprotID: Q8WZ42 | All beta proteins | 130 | 10
| 2.50e+02 | 138344000
| N/A | 4.39 | 1100 | 0.0055 | 20 | buffer 50 mM Tris HCl pH 8.0, 50 mM NaCl | 30000 | 1-30000 |  | Homo sapiens | No | IMMUNOGLOBULIN-LIKE DOMAIN | Optical Tweezers (OT) | constant force | DOI | 1996-07-11 | Improta, S., Politou, A.S., Pastore, A. | L Tskhovrebova, J Trinick, J A Sleep, R M Simmons - 1997 | | N_U[1100] |  | | |
| Barnase | 1BNI | Alpha and beta proteins (a+b) | 269 | 40
| 1.00e+09 |
| N/A | 10.2 | 38 | 3.37E-5 | N/A | ENCAD force field described in M. Levitt et al.1995, simulation at two pulling speeds: low rate where pulling speed= 0.01 angstrom/ps and high rate where pulling speed=0.02 angtrom/ps | 110 | 1-110 |  | Bacillus amyloliquefaciens | No | MICROBIAL RIBONUCLEASE | all atom SMD simulation | constant velocity | DOI | 1995-07-31 | Bycroft, M. | Robert B. Best, Bin Li, Annette Steward, Valerie Daggett, and Jane Clarke - 2001 | N/A | N_U[38] |  | | |
| Barnase linked to I27 | 1BNR: BARNASE | 1BNR | Alpha and beta proteins (a+b) | 70 |
| 1.00e+02 | 30
| N/A | 9.8 | 38 | 3.37E-5 | N/A | PBS: 10 mM phosphate buffer, 137 mM NaCl, 2.7 mM KCl), with the addition of 0.01% sodium azide to prevent bacterial and fungal growth | 110 | 1-110 |  | Bacillus amyloliquefaciens | No | MICROBIAL RIBONUCLEASE | AFM | constant velocity | DOI | 1995-07-31 | Bycroft, M. | Robert B. Best, Bin Li, Annette Steward, Valerie Daggett, and Jane Clarke - 2001 | N/A | N_U[38] |  | | |
| mutant C426A/D595A of CnaB domain and TED from Pilus-presented adhesin, Spy0125 (Cpa), P212121 form (DLS) | 2XID | Mainly Beta | 350 | 46
| 4.00e+02 | 100
| N/A | N/A | 107 | N/A | N/A | pH 7.2 with 10 m M Hepes, 150 m M NaCl, and 1 m M EDTA | 307 | 290-597 | 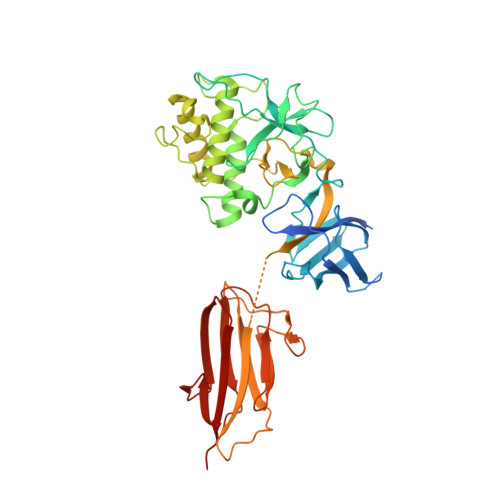 | Streptococcus pyogenes | yes | CELL ADHESION | AFM | constant force | DOI | 2010-08-04 | Pointon, J.A., Smith, W.D., Saalbach, G., Crow, A., Kehoe, M.A., Banfield, M.J. | Daniel J. Echelman, Alex Q. Lee, Julio M. Fernandez - 2017 | | N_I[55]_U[107] |  | 8.0E-5 | |
| 1st domain of synaptotagmin I (C2A) | 1DQV | All beta proteins | 60 |
| 6.00e+02 |
| N/A | N/A | 38 | N/A | N/A | 0.1 M imidazole PH 6.0 | 128 | 295-423 | 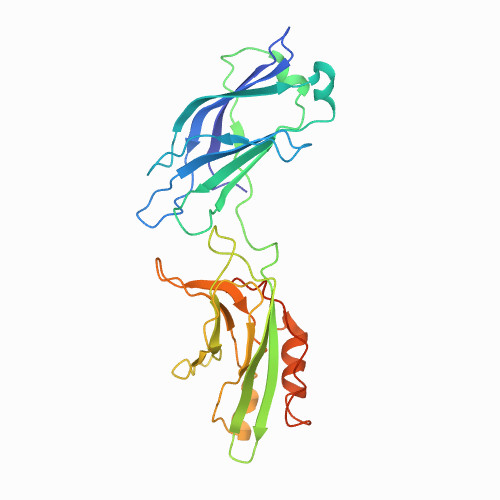 | Rattus rattus | No | ENDOCYTOSIS/EXOCYTOSIS | AFM | constant velocity | DOI | 2000-01-19 | Sutton, R.B., Ernst, J.A., Brunger, A.T. | Mariano Carrion-Vazquez*, Andres F. Oberhauser, Thomas E. Fisher, Piotr E. Marszalek, Hongbin Li, Julio M. Fernandez - 2000 | [A-G] | N_U[38] |  | | |
| domain 4 of ddFLN | 1WLH | All beta proteins | 75 | 1
| 2.00e+02 |
| N/A | N/A | 16.9 | 1.0E-5 | N/A | PDB and room temperature | 100 | 547-647 |  | Dictyostelium discoideum | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2004-10-05 | Popowicz, G.M., Mueller, R., Noegel, A.A., Schleicher, M., Huber, R., Holak, T.A. | Hendrik Dietz and Matthias Rief - 2004 | | N_I[15]_U[16] |  | | |
| Domain 12 of Fibronectin | 1FNF | All beta proteins | 125 | 18
| 4.00e+02 |
| N/A | N/A | 31 | N/A | 20 | PBS containing 5 mM DTT and 0.2 mM EDTA | 92 | 1833-1925 |  | Homo sapiens | No | HEPARIN AND INTEGRIN BINDING | AFM | constant velocity | DOI | 1999-03-16 | Sharma, A., Askari, J., Humphries, M., Jones, E.Y., Stuart, D.I. | Andres F Oberhauser, Carmelu Badilla-Fernandez, Mariano Carrion-Vazquez, Julio M Fernandez - 2002 | | |  | | |
| Cadherin-23 EC1-27 and Protocadherin-15 EC1-2 (handshake complex) in 'partner-assisted pulling-PAP ' mode | 4AQ8 | All beta proteins | 241 |
| 2.00e+03 | 0
| N/A | 18 | 33.2 | N/A | N/A | 10 mM HEPES, 50 mM KCl, 10 mM NaCl, and 50 µM CaCl 2 | | 1-210 | 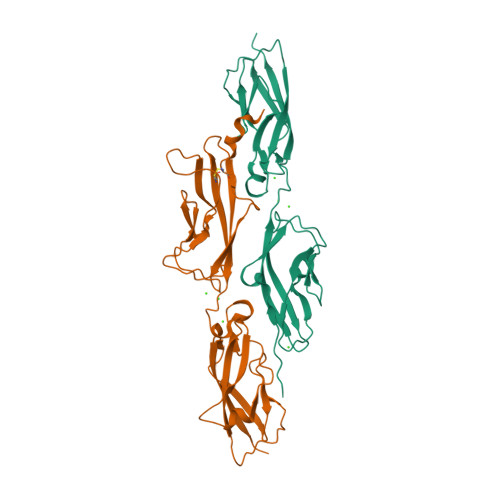 | Mus musculus | No | CELL ADHESION | AFM | constant velocity | DOI | 2012-11-07 | Sotomayor, M., Weihofen, W., Gaudet, R., Corey, D.P. | Nisha Arora, Jagadish Prasad Hazra, and Sabyasachi Rakshit - 2021 | | N_I[6.5]_I[16.9]_I[24.6]_U[33.2] |  | | |
| calmodulin | FREE CALMODULIN | 1CFC | All alpha proteins | 0 |
| 6.00e+02 |
| N/A | N/A | 194 | N/A | N/A | 0.1 M Tris PH 7.5 | 148 | 1-148 | 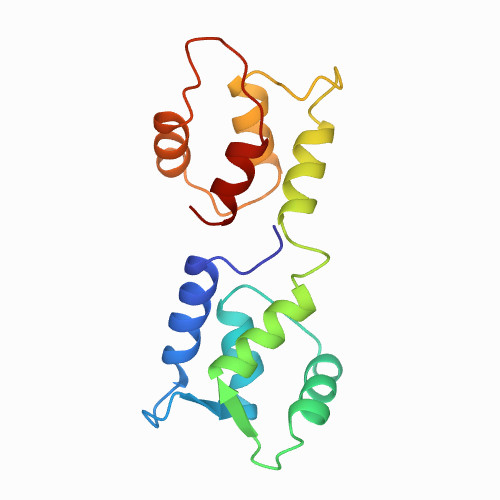 | Xenopus laevis | No | CALCIUM-BINDING PROTEIN | AFM | constant velocity | DOI | 1995-12-07 | Kuboniwa, H., Tjandra, N., Grzesiek, S., Ren, H., Klee, C.B., Bax, A. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2000 | No unfolding peaks: force below 20pN | N/A |  | | |
| construct of 6 Ankyrin repeats(NI6C) and polyprotein I27 of titin :(I27)(I27)(I27)-NI6C | 1N11 | Alpha and beta proteins (a+b) | 60 | 20
| 5.00e+02 |
| N/A | N/A | | N/A | N/A | N/A | 253 | 405-812 | 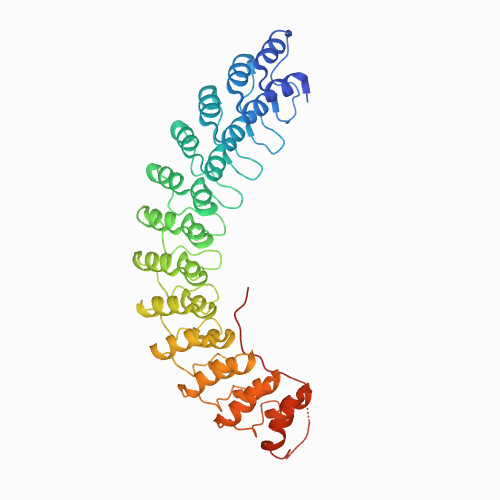 | Homo sapiens | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2002-12-11 | Michaely, P., Tomchick, D.R., Machius, M., Anderson, R.G.W. | Whasil Lee, Xiancheng Zeng, Kristina Rotolo, Ming Yang, Christopher J. Schofield, Vann Bennett, Weitao Yang, and Piotr E. Marszalek - 2012 | | N_U[8.8] |  | | |
| 6 repeats of Ankyrin (N6C) ; from second to seventh repeat | 1N11 | Alpha and beta proteins (a+b) | 37 | 20
| 5.00e+02 |
| N/A | N/A | 11.5 | N/A | 20 | N/A | 33 | 405-812 |  | Homo sapiens | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2002-12-11 | Michaely, P., Tomchick, D.R., Machius, M., Anderson, R.G.W. | Lewyn Li, Svava Wetzel, Andreas Pluckthun, Julio M. Fernandez - 2005 | | N_U[11.5] |  | | |
| lipoyl domain of the dihydrolipoyl acetyltransferase subunit from Escherichia coli (E2lip3(-)): pulling from C-ter to N-ter | 1QJO | All beta proteins | 177 | 3
| 7.00e+02 |
| N/A | N/A | 24.2 | 0.0076 | 20 | different directions of pulling: here from C-ter to N-ter | 79 | 1-80 | 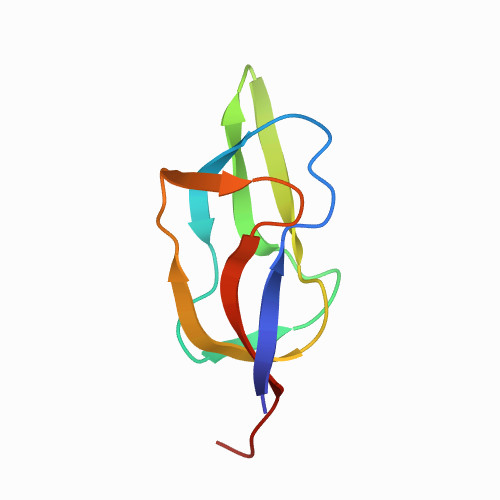 | Escherichia coli BL21(DE3) | No | TRANSFERASE | AFM | constant velocity | DOI | 1999-06-30 | Jones, D.D., Howard, M.J., Perham, R.N. | David J Brockwell, Emanuele Paci, Rebecca C Zinober, Godfrey S Beddard, Peter D Olmsted, D Alastair Smith, Richard N Perham, Sheena E Redford - 2003 | | N_U[24.2] |  | | |
| mutant C426A/D595A of CnaB domain and TED from Pilus-presented adhesin, Spy0125 (Cpa), P212121 form (DLS) | 2XID | Mainly Beta | 254 | 46
| 4.00e+02 |
| N/A | N/A | 119 | N/A | N/A | pH 7.2 with 10 m M Hepes, 150 m M NaCl, and 1 m M EDTA | 307 | 290-597 |  | Streptococcus pyogenes | yes | CELL ADHESION | AFM | constant force | DOI | 2010-08-04 | Pointon, J.A., Smith, W.D., Saalbach, G., Crow, A., Kehoe, M.A., Banfield, M.J. | Daniel J. Echelman, Alex Q. Lee, Julio M. Fernandez - 2017 | | N_U[119] |  | | |
| SdrG B1 in the absence of Calicum | 1R17 | All beta proteins | 650 |
| 3.40e+03 | 0
| 0.071 | N/A | 36 | 0.0082 | N/A | 10 mM of EDTA (No Calcium) | 320 | 276-596 |  | Staphylococcus epidermidis | No | CELL ADHESION | AFM | constant velocity | DOI | 2003-10-28 | Ponnuraj, K., Bowden, M.G., Davis, S., Gurusiddappa, S., Moore, D., Choe, D., Xu, Y., Hook, M., Narayana, S.V.L. | Lukas F. Milles, Eduard M. Unterauer1 , Thomas Nicolaus1 and Hermann E. Gaub - 2018 | | N_U[36] |  | | |
| SdrG B1 in presence of Calcium | 1R17 | All beta proteins | 2000 |
| 3.40e+03 | 317500
| 0.083 | N/A | 36 | 0.0082 | N/A | 10 mM of Calcium | 320 | 276-596 |  | Staphylococcus epidermidis | No | CELL ADHESION | AFM | constant velocity | DOI | 2003-10-28 | Ponnuraj, K., Bowden, M.G., Davis, S., Gurusiddappa, S., Moore, D., Choe, D., Xu, Y., Hook, M., Narayana, S.V.L. | Lukas F. Milles, Eduard M. Unterauer1 , Thomas Nicolaus1 and Hermann E. Gaub - 2018 | | N_U[36] |  | | |
| Cadherin-23 EC1-27 and Protocadherin-15 EC1-2 (handshake complex) in 'handle (or hook)-assisred pulling (HAP)' mode | 4AQ8 | All beta proteins | 195 |
| 2.00e+03 |
| 0.088 | 17 | 33.7 | N/A | N/A | handle (or hook)-assisred pulling (HAP) mode, refer to figure (by clicking on protein's name), buffer :10 mM HEPES, 50 mM KCl, 10 mM NaCl, and 50 uM CaCl 2 | | 1-210 |  | Mus musculus | No | CELL ADHESION | AFM | constant velocity | DOI | 2012-11-07 | Sotomayor, M., Weihofen, W., Gaudet, R., Corey, D.P. | Nisha Arora, Jagadish Prasad Hazra, and Sabyasachi Rakshit - 2021 | | N_I[6.4]_I[15.9]_I[24.5]_U[33.7] |  | | |
| Rubredoxin | 1BRF | Small proteins | 200 | 40
| 4.00e+02 | 0
| 0.11 | N/A | 13 | 0.15 | N/A | room temperature in Tris buffer at pH 7.4 | 53 | 1-53 | 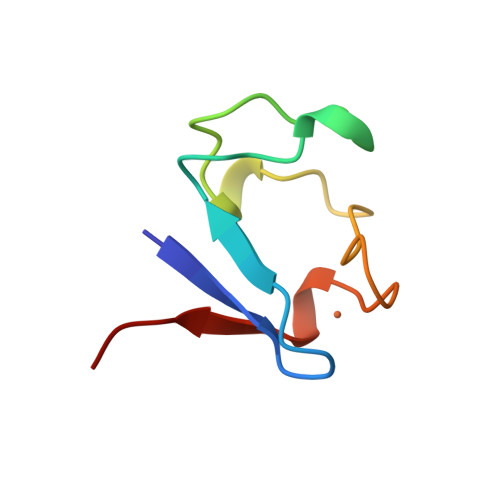 | Pyrococcus furiosus | No | ELECTRON TRANSPORT | AFM | constant velocity | DOI | 1998-09-02 | Bau, R., Rees, D.C., Kurtz, D.M., Scott, R.A., Huang, H., Adams, M.W.W., Eidsness, M.K. | Peng Zheng and Hongbin Li - 2011 | | N_U[13] |  | | |
| GFP pulling from residue 117 and residue 182 | 1EMB: GREEN FLUORESCENT PROTEIN (GFP) FROM AEQUOREA VICTORIA, GLN 80 REPLACED WITH ARG | 1EMB | Alpha and beta proteins (a+b) | 548 | 57
| 3.60e+03 |
| 0.12 | 31 | 72 | 5.0E-5 | N/A | PBS buffer (pH 7.4) at high protein concentrations of ~5 mg/ml (~0.2 mM) - Different pulling directions: see more in name column | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz, Felix Berkemeier, Morten Bertz, and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| GFP pulling from residue 3 and residue 132 | 1EMB: GREEN FLUORESCENT PROTEIN (GFP) FROM AEQUOREA VICTORIA, GLN 80 REPLACED WITH ARG | 1EMB | Alpha and beta proteins (a+b) | 407 | 45
| 12000 |
| 0.125 | 26 | 72 | 0.004 | N/A | PBS buffer (pH 7.4) at high protein concentrations of ~5 mg/ml (~0.2 mM) - Different pulling directions: see more in name column | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz, Felix Berkemeier, Morten Bertz, and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| scaffoldin c7A | 1AOH: SINGLE COHESIN DOMAIN FROM THE SCAFFOLDING PROTEIN CIPA OF THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME | 1AOH | All beta proteins | 1360 | 50
| 1.00e+12 | 100
| 0.131 | 21.9 | 49.3 | 6.0E-6 | 20 | CHARMM22 force fiels, water box 10 angstroms larger than the domain, neutralized by adding NaCl ions to the
water box, minimization was done with a time-step of 1 fs | 147 | 5-147 |  | Acetivibrio thermocellus ATCC 27405 | No | STRUCTURAL PROTEIN | all atom SMD simulation | constant velocity | DOI | 1998-07-08 | Alzari, P.M., Tavares, G. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][ A'-I '] | N_U[49.3] |  | | |
| scaffoldin c7A | 1AOH: SINGLE COHESIN DOMAIN FROM THE SCAFFOLDING PROTEIN CIPA OF THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME | 1AOH | All beta proteins | 710 | 50
| 1.00e+11 | 100
| 0.131 | 21.9 | 49.3 | 6.0E-6 | 20 | CHARMM22 force fiels, water box 10 angstroms larger than the domain, neutralized by adding NaCl ions to the
water box, minimization was done with a time-step of 1 fs | 147 | 5-147 |  | Acetivibrio thermocellus ATCC 27405 | No | STRUCTURAL PROTEIN | all atom SMD simulation | constant velocity | DOI | 1998-07-08 | Alzari, P.M., Tavares, G. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][ A'-I '] | N_U[49.3] |  | | |
| scaffoldin c7A | 1AOH: SINGLE COHESIN DOMAIN FROM THE SCAFFOLDING PROTEIN CIPA OF THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME | 1AOH | All beta proteins | 480 | 14
| 4.00e+02 | 100
| 0.131 | 21.9 | 49.3 | 6.0E-6 | 20 | PBS 0.2 mM EDTA 5mM DTT buffer | 147 | 5-147 |  | Acetivibrio thermocellus ATCC 27405 | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 1998-07-08 | Alzari, P.M., Tavares, G. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][ A'-I '] | N_U[49.3] |  | | |
| scaffoldin c1C | 1G1K: COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | 1G1K | All beta proteins | 425 | 9
| 4.00e+02 | 100
| 0.133 | 22.3 | 48.5 | 0.0002 | 20 | PBS 0.2 mM EDTA 5 mM DTT buffer | 143 | 2-143 | 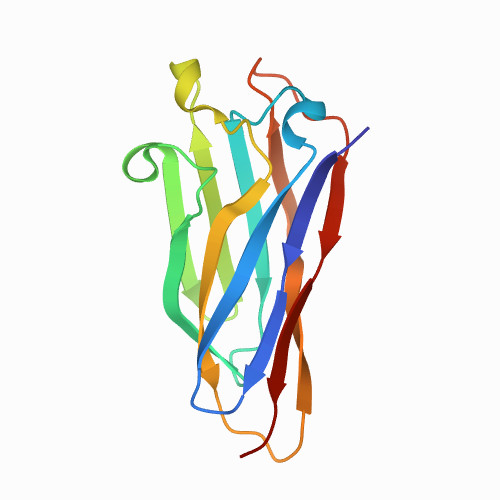 | Ruminiclostridium cellulolyticum | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2000-11-22 | Spinelli, S., Fierobe, H.-P., Belaich, A., Belaich, J.-P., Henrissat, B., Cambillau, C. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][A'-I'] | N_U[48.5] |  | | |
| scaffoldin c1C | 1G1K: COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | 1G1K | All beta proteins | 630 | 70
| 1.00e+09 | 100
| 0.133 | 22.3 | 48.5 | 0.0002 | 20 | CHARMM22 force fiels, water box 10 angstroms larger than the domain, neutralized by adding NaCl ions to the
water box, minimization was done with a time-step of 1 fs | 143 | 2-143 |  | Ruminiclostridium cellulolyticum | No | STRUCTURAL PROTEIN | all atom SMD simulation | constant velocity | DOI | 2000-11-22 | Spinelli, S., Fierobe, H.-P., Belaich, A., Belaich, J.-P., Henrissat, B., Cambillau, C. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][A'-I'] | N_U[48.5] |  | | |
| scaffoldin c1C | 1G1K: COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | 1G1K | All beta proteins | 1610 | 9
| 1.00e+10 |
| 0.133 | 22.3 | 48.5 | 0.0002 | 20 | CHARMM22 force fiels, water box 10 angstroms larger than the domain, neutralized by adding NaCl ions to the
water box, minimization was done with a time-step of 1 fs | 143 | 2-143 |  | Ruminiclostridium cellulolyticum | No | STRUCTURAL PROTEIN | all atom SMD simulation | constant velocity | DOI | 2000-11-22 | Spinelli, S., Fierobe, H.-P., Belaich, A., Belaich, J.-P., Henrissat, B., Cambillau, C | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][A'-I'] | N_U[48.5] |  | | |
| GFP pulling from residue 182 and residue 212 | 1EMB: GREEN FLUORESCENT PROTEIN (GFP) FROM AEQUOREA VICTORIA, GLN 80 REPLACED WITH ARG | 1EMB | Alpha and beta proteins (a+b) | 356 | 61
| 3.60e+03 |
| 0.14 | 27 | 72 | 0.0022 | N/A | PBS buffer (pH 7.4) at high protein concentrations of ~5 mg/ml (~0.2 mM) - Different pulling directions: see more in name column | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz, Felix Berkemeier, Morten Bertz, and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| Protein G domain B1 in the presence of glycerol | 1PGA | Alpha and beta proteins (a+b) | 250 | 50
| 1.00e+03 |
| 0.153 | 1.84 | 16.8 | 0.0059 | N/A | 10 mM Tris/HCl and 30% v/v glycerol (pH 7.5) | 56 | 1-56 | 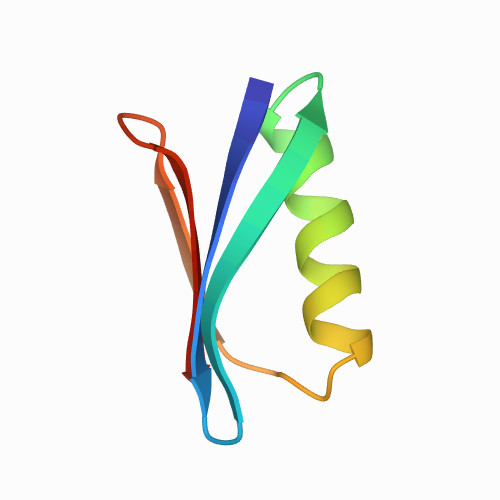 | Streptococcus sp. GX7805 | No | IMMUNOGLOBULIN BINDING PROTEIN | AFM | constant velocity | DOI | 1994-04-30 | Gallagher, T., Alexander, P., Bryan, P., Gilliland, G.L. | Daniel Aioanei, Isabella Tessari, Luigi Bubacco, Bruno Samori, Marco Brucale - 2011 | | N_U[16.8] |  | | |
| Protein G domain B1 in the absence of glycerol | 1PGA | Alpha and beta proteins (a+b) | 190 | 50
| 1.00e+03 |
| 0.165 | 0.84 | 16.8 | 0.0372 | N/A | Tris/HCl buffer (10 mM, pH 7.5) | 56 | 1-56 |  | Streptococcus sp. GX7805 | No | IMMUNOGLOBULIN BINDING PROTEIN | AFM | constant velocity | DOI | 1994-04-30 | Gallagher, T., Alexander, P., Bryan, P., Gilliland, G.L. | Daniel Aioanei, Isabella Tessari, Luigi Bubacco, Bruno Samori, Marco Brucale - 2011 | | N_U[16.8] |  | | |
| I27 of titin (also known as I91) | 1TIT | All beta proteins | 370 | 80
| 1.00e+11 |
| 0.168 | 17.3 | 27.3 | 0.0055 | 20 | CHARMM22 force fiels, water box 10 angstroms larger than the domain, neutralized by adding NaCl ions to the
water box, minimization was done with a time-step of 1 fs | 89 | 1-89 | 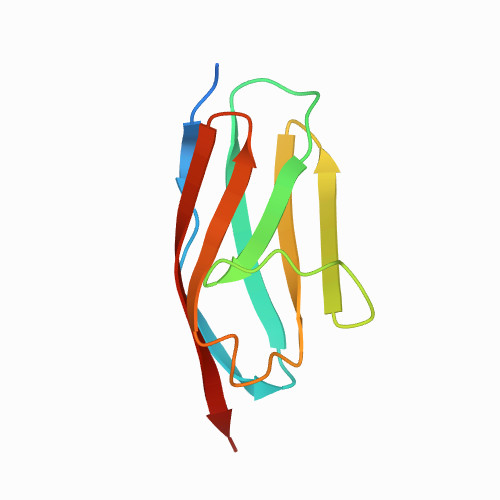 | Homo sapiens | No | IMMUNOGLOBULIN-LIKE DOMAIN | all atom SMD simulation | constant velocity | DOI | 1996-07-11 | Improta, S., Politou, A.S., Pastore, A. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-B][A'-G] | N_U[27.3] |  | | |
| I27 of titin (also known as I91) | 1TIT | All beta proteins | 910 | 50
| 1.00e+12 |
| 0.168 | 17.3 | 27.3 | 0.0055 | 20 | CHARMM22 force fiels, water box 10 angstroms larger than the domain, neutralized by adding NaCl ions to the
water box, minimization was done with a time-step of 1 fs | 89 | 1-89 |  | Homo sapiens | No | IMMUNOGLOBULIN-LIKE DOMAIN | all atom SMD simulation | constant velocity | DOI | 1996-07-11 | Improta, S., Politou, A.S., Pastore, A. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-B][A'-G] | N_U[27.3] |  | | |
| I27 of titin (also known as I91) | 1TIT | All beta proteins | 211 | 1
| 1.00e+03 |
| 0.168 | 17.3 | 27.3 | 0.0055 | 20 | PBS 0.2 mM EDTA 5mM DTT buffer | 89 | 1-89 |  | Homo sapiens | No | IMMUNOGLOBULIN-LIKE DOMAIN | AFM | constant velocity | DOI | 1996-07-11 | Improta, S., Politou, A.S., Pastore, A. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-B][A'-G] | N_U[27.3] |  | | |
| 2nd Domain of Fibronection III (FNIII) | 1OWW | All beta proteins | 220 | 18
| 6.00e+02 |
| 0.17 | 0.004 | 31 | 0.022 | 20 | PBS containing 5 mM DTT and 0.2 mM EDTA | 91 | 739-830 |  | Homo sapiens | No | CELL ADHESION PROTEIN | AFM | constant velocity | DOI | 1996-01-29 | Leahy, D.J., Aukhil, I., Erickson, H.P. | Lewyn Li 1, Hector Han-Li Huang, Carmen L Badilla, Julio M Fernandez - 2004 | [A-B][A-G] | N_U[31] |  | | |
| scaffoldin c2A | Cohesin 2 DOMAIN OF THE CELLULOSOME FROM CLOSTRIDIUM THERMOCELLUM | 1ANU | All beta proteins | 860 | 50
| 1.00e+12 |
| 0.17 | 18.4 | 48.6 | 0.021 | 20 | CHARMM22 force fiels, water box 10 angstroms larger than the domain, neutralized by adding NaCl ions to the
water box, minimization was done with a time-step of 1 fs | 138 | 1-138 |  | Acetivibrio thermocellus | No | COHESIN | all atom SMD simulation | constant velocity | DOI | 1997-07-23 | Shimon, L.J.W., Yaron, S., Shoham, Y., Lamed, R., Morag, E., Bayer, E.A., Frolow, F. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][ A'-I '] | N_U[48.6] |  | | |
| 1st domain of Fibronectin III (FNIII) | 1FNH | All beta proteins | 200 | 18
| 6.00e+02 |
| 0.17 | 20.5 | 28 | 0.022 | 20 | PBS containing 5 mM DTT and 0.2 mM EDTA | 97 | 625-722 |  | Homo sapiens | No | CELL ADHESION PROTEIN | AFM | constant velocity | DOI | 1996-01-29 | Leahy, D.J., Aukhil, I., Erickson, H.P. | Andres F Oberhauser, Carmelu Badilla-Fernandez, Mariano Carrion-Vazquez, Julio M Fernandez - 2002 | [A-B][A-G] | N_I[20]_U[28] |  | | |
| scaffoldin c2A | Cohesin 2 DOMAIN OF THE CELLULOSOME FROM CLOSTRIDIUM THERMOCELLUM | 1ANU | All beta proteins | 470 | 80
| 1.00e+11 |
| 0.17 | 18.4 | 48.6 | 0.021 | 20 | CHARMM22 force fiels, water box 10 angstroms larger than the domain, neutralized by adding NaCl ions to the
water box, minimization was done with a time-step of 1 fs | 138 | 1-138 |  | Acetivibrio thermocellus | No | COHESIN | all atom SMD simulation | constant velocity | DOI | 1997-07-23 | Shimon, L.J.W., Yaron, S., Shoham, Y., Lamed, R., Morag, E., Bayer, E.A., Frolow, F. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][ A'-I '] | N_U[48.6] |  | | |
| scaffoldin c2A | Cohesin 2 DOMAIN OF THE CELLULOSOME FROM CLOSTRIDIUM THERMOCELLUM | 1ANU | All beta proteins | 214 | 8
| 4.00e+02 | 100
| 0.17 | 18.4 | 48.6 | 0.021 | 20 | PBS 0.2 mM EDTA 5 mM DTT buffer | 138 | 1-138 |  | Acetivibrio thermocellus | No | COHESIN | AFM | constant velocity | DOI | 1997-07-23 | Shimon, L.J.W., Yaron, S., Shoham, Y., Lamed, R., Morag, E., Bayer, E.A., Frolow, F. | Alejandro Valbuena, Javier Oroz, Ruben Hervas, and Mariano Carrion-Vazquez - 2009 | [A-I][ A'-I '] | N_U[48.6] |  | | |
| protein L | 1HZ6: CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | 1HZ6 | Alpha and beta proteins (a+b) | 500 | 200
| 4.00e+09 |
| 0.22 | N/A | 18.8 | 0.05 | 20 | N/A | 62 | 2-64 |  | Finegoldia magna ATCC 29328 | Yes | PROTEIN BINDING | all atom SMD simulation | constant velocity | DOI | 2001-04-04 | O'Neill, J.W., Kim, D.E., Baker, D., Zhang, K.Y.J. | David J. Brockwell, Godfrey S. Beddard, Emanuele Paci, Peter D. Olmsted, D. Alastair Smith, and Sheena E Redford - 2005 | | N_U[18.8] |  | | |
| protein L | 1HZ6: CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | 1HZ6 | Alpha and beta proteins (a+b) | 91 | 3
| 40 |
| 0.22 | N/A | 18.8 | 0.05 | 20 | PBS | 62 | 2-64 |  | Finegoldia magna ATCC 29328 | Yes | PROTEIN BINDING | AFM | constant velocity | DOI | 2001-04-04 | O'Neill, J.W., Kim, D.E., Baker, D., Zhang, K.Y.J. | David J. Brockwell, Godfrey S. Beddard, Emanuele Paci, Peter D. Olmsted, D. Alastair Smith, and Sheena E Redford - 2005 | | N_U[18.8] |  | | |
| Native Ubiquitin at PH=2 | 1UBQ | All alpha proteins | 180 | 34
| 1.00e+03 |
| 0.225 | N/A | 24.5 | 0.02 | N/A | forces were measured at different PH, here PH=2. buffer PBS: 126 mM NaCl, 7.2 mM Na2HPO4, 3 mM NaH2PO4 | | 1-76 | 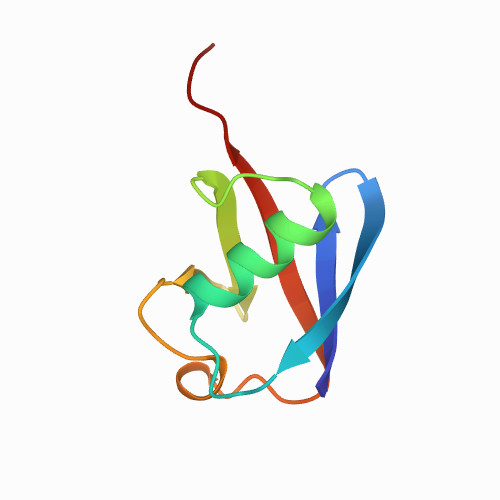 | Gallus gallus | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2004-10-19 | Kusunoki, H., Minasov, G., MacDonald, R.I., Mondragon, A. | Chia-Lin Chyan, Fan-Chi Lin, Haibo Peng, Jian-Min Yuan, Chung-Hung Chang, Sheng-Hsien Lin, and Guoliang Yang - 2004 | | N_U[24] |  | | |
| Native Ubiquitin at PH from 6 to 10 | 1UBQ | All alpha proteins | 230 | 34
| 1000 |
| 0.225 | N/A | 24.5 | 5.0E-5 | N/A | forces were measured at different PH, here PH from 6 to 10. buffer PBS: 126 mM NaCl, 7.2 mM Na2HPO4, 3 mM NaH2PO4 | | 1-76 |  | Gallus gallus | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2004-10-19 | Kusunoki, H., Minasov, G., MacDonald, R.I., Mondragon, A. | Chia-Lin Chyan, Fan-Chi Lin, Haibo Peng, Jian-Min Yuan, Chung-Hung Chang, Sheng-Hsien Lin, and Guoliang Yang - 2004 | | N_U[24] |  | | |
| N-C linked ubiquitin | 1UBQ | Alpha and beta proteins (a+b) | 2000 | 35
| 1.00e+11 |
| 0.25 | N/A | 24 | 0.0004 | N/A | CHARMM22 force field, NPT ensemble at contant velocity, periodic boundary condition | 76 | 1-76 |  | Homo sapiens | No | CHROMOSOMAL PROTEIN | all atom SMD simulation | constant velocity | DOI | 1987-04-16 | Vijay-Kumar, S., Bugg, C.E., Cook, W.J. | Mariano Carrion-Vazquez 1 , Hongbin Li, Hui Lu, Piotr E Marszalek, Andres F Oberhauser, Julio M Fernandez - 2003 | | N_U[24] |  | | |
| I27 of titin (also known as I91) | 1TIT | All beta proteins | 225 | 1
| 1000 |
| 0.25 | N/A | 28.7 | 0.0055 | N/A | 25 mM imidazole-HCl, pH 7.4, 0.2M KCI,4mM | 89 | 1-89 |  | Homo sapiens | No | IMMUNOGLOBULIN-LIKE DOMAIN | AFM | constant velocity | DOI | 1996-07-11 | Improta, S., Politou, A.S., Pastore, A. | Kaori Watanabe, Claudia Muhle-Goll, Miklos S Z Kellermayer, Siegfried Labeit, Henk Granzier - 2002 | [A-B][A'-G] | N_U[28.7] |  | | |
| N-C linked ubiquitin | 1UBQ | Alpha and beta proteins (a+b) | 203 | 35
| 4.00e+02 |
| 0.25 | N/A | 24 | 0.0004 | N/A | examination of the mechanical stability of N-C linked. buffer: 50 mM Tris, pH 7.6 | 76 | 1-76 |  | Homo sapiens | No | CHROMOSOMAL PROTEIN | AFM | constant velocity | DOI | 1987-04-16 | Vijay-Kumar, S., Bugg, C.E., Cook, W.J. | Mariano Carrion-Vazquez, Hongbin Li, Hui Lu, Piotr E Marszalek, Andres F Oberhauser, Julio M Fernandez - 2003 | | N_U[24] |  | | |
| lipoyl domain of the dihydrolipoyl acetyltransferase subunit from Escherichia coli (E2lip3(+)): pulling from C-ter to Residue 41 (Lys) | 1QJO | All beta proteins | 187 | 3
| 7.00e+02 |
| 0.29 | N/A | 24.2 | 0.0076 | 20 | different directions of pulling: here from C-ter to Lys 41 | 79 | 1-80 |  | Escherichia coli BL21(DE3) | No | TRANSFERASE | AFM | constant velocity | DOI | 1999-06-30 | Jones, D.D., Howard, M.J., Perham, R.N. | David J Brockwell, Emanuele Paci, Rebecca C Zinober, Godfrey S Beddard, Peter D Olmsted, D Alastair Smith, Richard N Perham, Sheena E Redford - 2003 | | N_I[10.9]_U[24.2] |  | | |
| GFP(132,212) :from copolymers with mixed linkage
geometries: (3, 212) and (132, 212)| 1EMB: GREEN FLUORESCENT PROTEIN (GFP) FROM AEQUOREA VICTORIA, GLN 80 REPLACED WITH ARG | 1EMB | Alpha and beta proteins (a+b) | 117 | 22
| 3.60e+03 |
| 0.32 | 25 | 72 | 0.014 | N/A | PBS buffer (pH 7.4) at high protein concentrations of ~5 mg/ml (~0.2 mM) - Different pulling directions: see more in name column | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| GFP polyprotein linked by cysteine on 132-212| 1EMB: GREEN FLUORESCENT PROTEIN (GFP) FROM AEQUOREA VICTORIA, GLN 80 REPLACED WITH ARG | 1EMB | Alpha and beta proteins (a+b) | 117 | 22
| 3.60e+03 |
| 0.32 | 25 | 72 | 0.014 | N/A | PBS buffer (pH 7.4) at high protein concentrations of ~5 mg/ml (~0.2 mM) - Different pulling directions: see more in name column | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz, Felix Berkemeier, Morten Bertz, and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| GFP pulling from residue 132 and residue 212 | 1EMB: GREEN FLUORESCENT PROTEIN (GFP) FROM AEQUOREA VICTORIA, GLN 80 REPLACED WITH ARG | 1EMB | Alpha and beta proteins (a+b) | 127 | 22
| 3.60e+03 |
| 0.32 | 26 | 72 | 0.005 | N/A | PBS buffer (pH 7.4) at high protein concentrations of ~5 mg/ml (~0.2 mM) - Different pulling directions: see more in name column | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz, Felix Berkemeier, Morten Bertz, and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| Domain 13 of Fibronectin III (FNIII) | 1FNF | All beta proteins | 89 | 18
| 6.00e+02 |
| 0.34 | 19.5 | 28 | 0.022 | 20 | PBS containing 5 mM DTT and 0.2 mM EDTA | 89 | 1926-2015 |  | Homo sapiens | No | CELL ADHESION PROTEIN | AFM | constant velocity | DOI | 1996-01-29 | Leahy, D.J., Aukhil, I., Erickson, H.P. | Andres F Oberhauser, Carmelu Badilla-Fernandez, Mariano Carrion-Vazquez, Julio M Fernandez - 2002 | [A-B][A-G] | N_U[31] |  | | |
| I1 DOMAIN OF TITIN | 1G1C | All beta proteins | 127 | 18
| 6.00e+02 |
| 0.35 | N/A | 30 | 0.005 | N/A | PBS buffer | 100 | 1-100 | 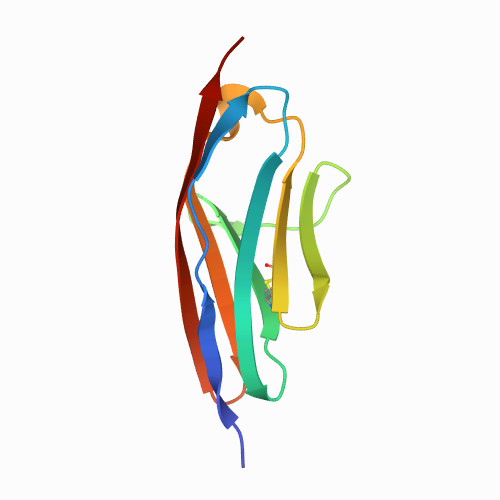 | Homo sapiens | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2001-10-12 | Mayans, O., Wuerges, J., Gautel, M., Wilmanns, M. | Hongbin Li, Julio M Fernandez - 2003 | [A-B A'-G] | N_U[30] |  | | |
| I65 to I70 Igs of titin | 3B43 | All beta proteins | 200 |
| 200 |
| 0.35 | N/A | 31.9 | 1.0E-5 | 20 | 25 mM imidazole-HCl, PH 7.4, 0.2 M KCl, 4mM MgCl2, 1mM EGTA, 0.01% NaN3, 5mM DTT | 572 | 1-573 | 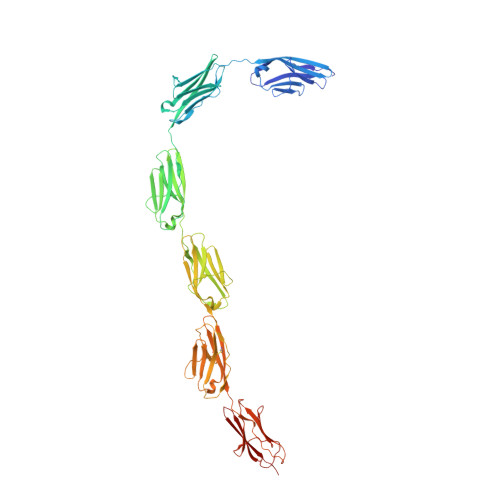 | Oryctolagus cuniculus | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2008-01-22 | von Castelmur, E., Marino, M., Labeit, D., Labeit, S., Mayans, O. | Kaori Watanabe, Claudia Muhle-Goll, Miklós S Z Kellermayer, Siegfried Labeit, Henk Granzier - 2002 | [A-B][A'-G] | N_U[31.9] |  | | |
| V17P mutant of I1 domain of TITIN | 1G1C | All beta proteins | 117 | 42
| 6.00e+02 |
| 0.35 | N/A | 30 | 0.008 | N/A | PBS buffer | 100 | 1-100 |  | Homo sapiens | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2001-10-12 | Mayans, O., Wuerges, J., Gautel, M., Wilmanns, M. | Hongbin Li, Julio M Fernandez - 2003 | [A-B A'-G] | N_U[30] |  | | |
| Q15P mutant of I1 domain of TITIN | 1G1C | All beta proteins | 82 | 28
| 6.00e+02 |
| 0.35 | N/A | 30 | 0.05 | N/A | PBS buffer | 100 | 1-100 |  | Homo sapiens | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2001-10-12 | Mayans, O., Wuerges, J., Gautel, M., Wilmanns, M. | Hongbin Li, Julio M Fernandez - 2003 | [A-B A'-G] | N_U[30] |  | | |
| R28P mutant of I1 domain of TITIN | 1G1C | All beta proteins | 82 | 28
| 6.00e+02 |
| 0.35 | N/A | 30 | 0.05 | N/A | PBS buffer | 100 | 1-100 |  | Homo sapiens | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2001-10-12 | Mayans, O., Wuerges, J., Gautel, M., Wilmanns, M. | Hongbin Li, Julio M Fernandez - 2003 | [A-B A'-G] | N_U[30] |  | | |
| E4P mutant domain I1 of TITIN | 1G1C | All beta proteins | 80 | 23
| 6.00e+02 |
| 0.35 | N/A | 30 | 0.07 | N/A | PBS buffer | 100 | 1-100 |  | Homo sapiens | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2001-10-12 | Mayans, O., Wuerges, J., Gautel, M., Wilmanns, M. | Hongbin Li, Julio M Fernandez - 2003 | [A-B A'-G] | N_U[30] |  | | |
| K7P mutant of domain I1 of TITIN | 1G1C | All beta proteins | 69 | 30
| 6.00e+02 |
| 0.35 | N/A | 30 | 0.1 | N/A | PBS buffer | 100 | 1-100 |  | Homo sapiens | No | STRUCTURAL PROTEIN | AFM | constant velocity | DOI | 2001-10-12 | Mayans, O., Wuerges, J., Gautel, M., Wilmanns, M. | Hongbin Li, Julio M Fernandez - 2003 | [A-B A'-G] | N_U[30] |  | | |
| One step unfolding of Domain 13 of Fibronectin III (FNIII) | 1FNF | All beta proteins | 100 | 10
| 3.00e+02 |
| 0.38 | N/A | 31 | 0.02 | 20 | buffer 50 mM Tris HCl pH 8.0, 50 mM NaCl | 94 | 1559-1653 |  | Homo sapiens | No | CELL ADHESION PROTEIN | AFM | constant velocity | DOI | 1996-01-29 | Leahy, D.J., Aukhil, I., Erickson, H.P. | Lewyn Li 1, Hector Han-Li Huang, Carmen L Badilla, Julio M Fernandez - 2004 | [A-B][A-G] | N_U[31] |  | | |
| Two steps unfolding of Domain 13 of Fibronectin III (FNIII) | 1FNF | All beta proteins | 75 |
| 4.00e+02 |
| 0.38 | N/A | 31 | 0.02 | 20 | PBS containing 5 mM DTT and 0.2 mM EDTA | 94 | 1559-1653 |  | Homo sapiens | No | CELL ADHESION PROTEIN | AFM | constant velocity | DOI | 1996-01-29 | Leahy, D.J., Aukhil, I., Erickson, H.P. | Lewyn Li, Hector Han-Li Huang, Carmen L. Badilla and Julio M. Fernandez - 2002 | [A-B][A-G] | N_I[12]_U[31] |  | | |
| WildType bacteriodepsin | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | 1AT9 | All alpha proteins | 150 | 50
| 4.00e+01 | 1000
| 0.4 | N/A | 40 | N/A | 20 | buffer solution (300 mM KCl, 10 mM tris-HCl, pH 7.8) | 231 | 1-231 |  | Halobacterium salinarum | No | PHOTORECEPTOR | AFM | constant velocity | DOI | 1998-09-16 | Kimura, Y., Vassylyev, D.G., Miyazawa, A., Kidera, A., Matsushima, M., Mitsuoka, K., Murata, K., Hirai, T., Fujiyoshi, Y. | F Oesterhelt, D Oesterhelt, M Pfeiffer, A Engel, H E Gaub, D J Muller - 2000 | [G-F & E-D][B][C] | N_I[10]_I|30]_I|50]_U[70] |  | | |
| bacteriorhodopsin muted by cleavage of E-F loop | 1AT9: STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | 1AT9 | All alpha proteins | 100 | 50
| 4.00e+01 | 1000
| 0.4 | N/A | 40 | N/A | 20 | buffer solution (300 mM KCl, 10 mM tris-HCl, pH 7.8) | 231 | 1-231 |  | Halobacterium salinarum | No | PHOTORECEPTOR | AFM | constant velocity | DOI | 1998-09-16 | Kimura, Y., Vassylyev, D.G., Miyazawa, A., Kidera, A., Matsushima, M., Mitsuoka, K., Murata, K., Hirai, T., Fujiyoshi, Y. | F Oesterhelt, D Oesterhelt, M Pfeiffer, A Engel, H E Gaub, D J Muller - 2000 | [D-E][B-C][A] | N_I[18]_I[26]_U[40] |  | | |
| G241C mutant of bacteriodepsin | 1AT9: STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | 1AT9 | All alpha proteins | 150 | 50
| 4.00e+01 | 1000
| 0.4 | N/A | 40 | N/A | 20 | buffer solution (300 mM KCl, 10 mM tris-HCl, pH 7.8) | 231 | 1-231 |  | Halobacterium salinarum | No | PHOTORECEPTOR | AFM | constant velocity | DOI | 1998-09-16 | Kimura, Y., Vassylyev, D.G., Miyazawa, A., Kidera, A., Matsushima, M., Mitsuoka, K., Murata, K., Hirai, T., Fujiyoshi, Y. | F Oesterhelt, D Oesterhelt, M Pfeiffer, A Engel, H E Gaub, D J Muller - 2000 | [F-G][D-E][B-C][A] | N_I[10]_I[25]_I[45]_U[65] |  | | |
| GFP(3-212) :from copolymers with mixed linkage
geometries: (3, 212) and (132, 212)| 1EMB: GREEN FLUORESCENT PROTEIN (GFP) FROM AEQUOREA VICTORIA, GLN 80 REPLACED WITH ARG | 1EMB | Alpha and beta proteins (a+b) | 115 | 22
| 3.60e+03 |
| 0.45 | 28 | 72 | 0.0008 | N/A | PBS buffer (pH 7.4) at high protein concentrations of ~5 mg/ml (~0.2 mM) - Different pulling directions: see more in name column | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| GFP polyprotein 2-212 | 1EMB | Alpha and beta proteins (a+b) | 117 | 22
| 1.20e+04 |
| 0.45 | 28 | 72 | 0.014 | N/A | Measured for GFP polyprotein linked by Cysteine on 3-212 and When pulling from residue 3(N-ter) & 212(C-ter) | 229 | 2-229 |  | Aequorea victoria | Yes | FLUORESCENT PROTEIN | AFM | constant velocity | DOI | 1997-06-16 | Brejc, K., Sixma, T. | Hendrik Dietz, Felix Berkemeier, Morten Bertz, and Matthias Rief - 2006 | N/A | N_I[39.3]_U[72.08] |  | | |
| The 3rd spectrin repeat of alpha-actinin-4 | 1WLX | All alpha proteins | 75 | 1
See more | 100000 | 1154.72
See more | 0.53 | 18.4 | 34 | 11 | N/A | PBS: 10 mM Na2HPO4 , 1.76 mM KH2 PO 4 , 137 mM NaCl, 2.7 mM KCl at room temperature | 129 | 1-129 |  | Homo sapiens | No | PROTEIN BINDING | High Speed AFM | constant velocity | DOI | 2005-08-09 | Kowalski, K., Merkel, A.L., Booker, G.W. | Hirohide Takahashi, Felix Rico, Christophe Chipot, and Simon Scheuring - 2018 | | N_I[10]_U[34] |  | | |
| GFP linked to eight Ig modules (Ig27 - Ig34) | 1B9C: Green Fluorescent Protein Mutant F99S, M153T and V163A | 1B9C | Alpha and beta proteins (a+b) | 104 | 40
| 3.00e+02 |
| 0.55 | 22 | 76.6 | N/A | 20 | 20 ul of protein solution (PBS buffer, pH 7.4; protein concentration, ~0.5 g/liter) | 227 | 4-230 |  | Aequorea victoria | Yes | LUMINESCENT PROTEIN | AFM | constant velocity | DOI | 2000-11-17 | Battistutta, R., Negro, A., Zanotti, G. | Hendrik Dietz and Matthias Rief - 2004 | [last alpha helix][last beta strand] | N_I[3.2]_I[0.28]_I[6.5]_I[0.55]_U[69.3] |  | | |
| GFP linked to DdFLN1-5 [actin crosslinker dictyostelium discoideum filamin] | 1B9C: Green Fluorescent Protein Mutant F99S, M153T and V163A | 1B9C | Alpha and beta proteins (a+b) | 104 | 40
| 3.00e+02 |
| 0.55 | 22 | 76.6 | N/A | 20 | 20 ul of protein solution (PBS buffer, pH 7.4; protein concentration, ~0.5 g/liter) | 227 | 4-230 |  | Aequorea victoria | Yes | LUMINESCENT PROTEIN | AFM | constant velocity | DOI | 2000-11-17 | Battistutta, R., Negro, A., Zanotti, G. | Hendrik Dietz and Matthias Rief - 2004 | [last alpha helix][last beta strand] | N_I[3.2]_I[0.28]_I[6.5]_I[0.55]_U[69.3] |  | | |
| Lys48-C linked poly-ubiquitin | 1UBQ | Alpha and beta proteins (a+b) | 1200 | 1
| 1.00e+11 |
| 0.63 | N/A | 7.8 | 0.0004 | N/A | CHARMM22 force field,NPT ensemble at contant velocity, periodic boundary condition | 76 | 1-76 |  | Homo sapiens | No | CHROMOSOMAL PROTEIN | all atom SMD simulation | constant velocity | DOI | 1987-04-16 | Vijay-Kumar, S., Bugg, C.E., Cook, W.J. | Mariano Carrion-Vazquez 1 , Hongbin Li, Hui Lu, Piotr E Marszalek, Andres F Oberhauser, Julio M Fernandez - 2003 | [strand3 - strand4] | N_U[7] |  | | |
| Lys48-C linked poly-ubiquitin | 1UBQ | Alpha and beta proteins (a+b) | 85 | 20
| 3.00e+02 |
| 0.63 | N/A | 7.8 | 0.0004 | N/A | examination of the mechanical stability of Lys48-C linked ubiquitin. buffer: 50 mM Tris, pH 7.6 | 76 | 1-76 |  | Homo sapiens | No | CHROMOSOMAL PROTEIN | AFM | constant velocity | DOI | 1987-04-16 | Vijay-Kumar, S., Bugg, C.E., Cook, W.J. | Mariano Carrion-Vazquez, Hongbin Li, Hui Lu, Piotr E Marszalek, Andres F Oberhauser, Julio M Fernandez - 2003 | [strand3 - strand4] | N_U[7] |  | | |
| T4 Lysozyme | 1B6I: T4 LYSOZYME MUTANT WITH CYS 54 REPLACED BY THR, CYS 97 REPLACED BY ALA, THR 21 REPLACED BY CYS AND LYS 124 REPLACED BY CYS (C54T,C97A,T21C,K124C) | 1B6I | Alpha and beta proteins (a+b) | 64 | 16
| 1.00e+03 |
| 0.81 | 7.6 | 36 | 0.0001 | 20 | PBS | 162 | 1-162 | 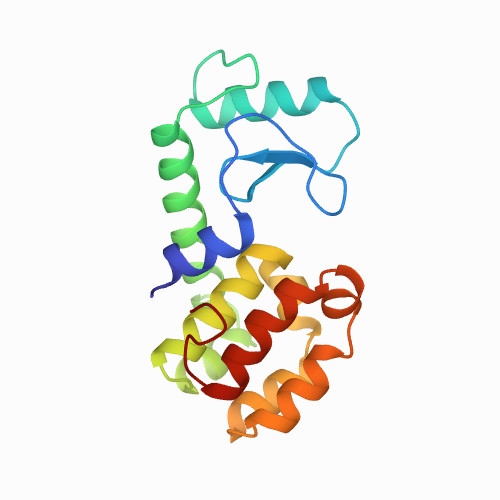 | Escherichia virus T4 | No | HYDROLASE | Scanning Force Microscope (SFM) | constant velocity | DOI | 2000-01-12 | Vetter, I.R., Baase, W.A., Snow, S., Matthews, B.W. | Guoliang Yang, Ciro Cecconi, Walter A. Baase, and Carlos Bustamante - 2000 | | |  | | |
| I27 of titin (also known as I91) | 1TIT | All beta proteins | 350 | 1
| 1000000 | 138344000
| 0.89 | 36 | | 2.0E-10 | N/A | buffer 50 mM Tris HCl pH 8.0, 50 mM NaCl | 89 | 1-89 |  | Homo sapiens | No | IMMUNOGLOBULIN-LIKE DOMAIN | High Speed AFM | constant velocity | DOI | 1996-07-11 | Improta, S., Politou, A.S., Pastore, A. | Felix Rico, Laura Gonzalez, Ignacio Casuso, Manel Puig-Vidal, and Simon Scheuring - 2013 | [A-B][A'-G] | |  | | |
| I27 of titin (also known as I91) | 1TIT | All beta proteins | 300 | 1
| 100000 | 44594100
| 0.89 | 36 | | 2.0E-10 | N/A | NPT ensemble, periodic boundary conditions, constant velocity pulling | 89 | 1-89 |  | Homo sapiens | No | IMMUNOGLOBULIN-LIKE DOMAIN | all atom SMD simulation | constant velocity | DOI | 1996-07-11 | Improta, S., Politou, A.S., Pastore, A. | Felix Rico, Laura Gonzalez, Ignacio Casuso, Manel Puig-Vidal, and Simon Scheuring - 2013 | [A-B][A'-G] | |  | | |
| Vascular Adhesion Molecule 1 (VCAM1) in presence of reducing agent | 1VSC | All alpha proteins | 40 | 1
| 1.00e+03 |
| 1.365 | N/A | | 0.0001 | 23 | at T = 23 C with reduction 1 mM TCEP | 196 | 1-196 |  | Mus musculus | No | STRUCTURAL GENOMICS, UNKNOWN FUNCTION | AFM | constant velocity | DOI | 2005-05-03 | Abe, T., Hirota, H., Tomizawa, T., Koshiba, S., Kigawa, T., Yokoyama, S., RIKEN Structural Genomics/Proteomics Initiative (RSGI) | Nishant Bhasin, Philippe Carl, Sandy Harper, Gang Feng, Hui Lu, David W. Speicher, Dennis E. Discher - 2004 | | |  | | |
| Native Vacsular Cellular Adhesion Molecule 1 (VCAM1) | 1VSC | All alpha proteins | 40 | 1
| 1.00e+03 |
| 1.6 | N/A | | 1.0E-7 | 23 | at T = 23 C and without any reducing agent | 196 | 1-196 |  | Mus musculus | No | STRUCTURAL GENOMICS, UNKNOWN FUNCTION | AFM | constant velocity | DOI | 2005-05-03 | Abe, T., Hirota, H., Tomizawa, T., Koshiba, S., Kigawa, T., Yokoyama, S., RIKEN Structural Genomics/Proteomics Initiative (RSGI) | Nishant Bhasin, Philippe Carl, Sandy Harper, Gang Feng, Hui Lu, David W. Speicher, Dennis E. Discher - 2004 | | |  | | |
| Spectrin | 1AJ3 | All alpha proteins | 30 | 5
| 3.00e+02 |
| 1.7 | N/A | 31.3 | 0.0003 | 20 | PBS | 106 | 1-106 |  | Gallus gallus | No | CYTOSKELETON | AFM | constant velocity | DOI | | Pascual, J., Pfuhl, M., Walther, D., Saraste, M., Nilges, M | M Rief, J Pascual, M Saraste, H E Gaub - 1999 | N/A | N_U[31.3] |  | | |