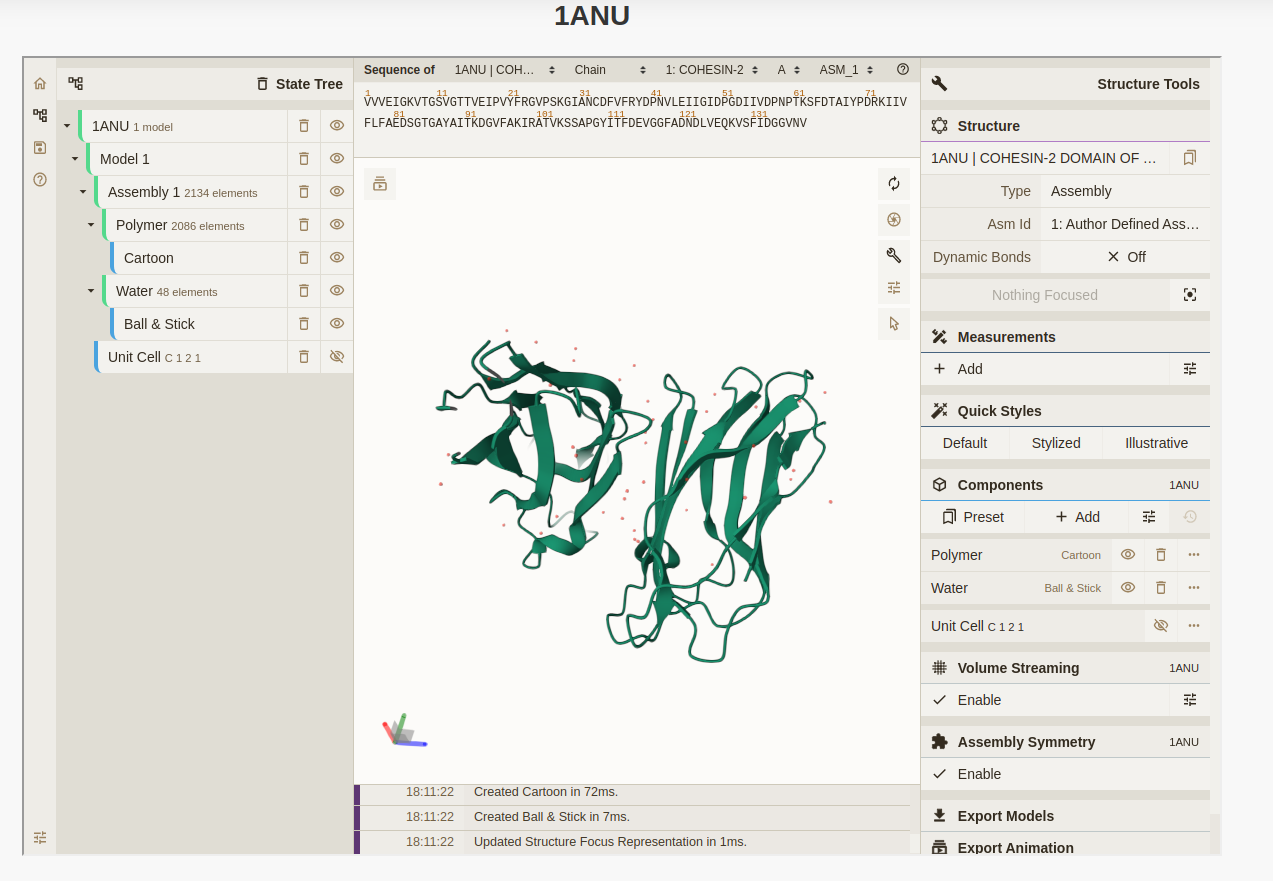
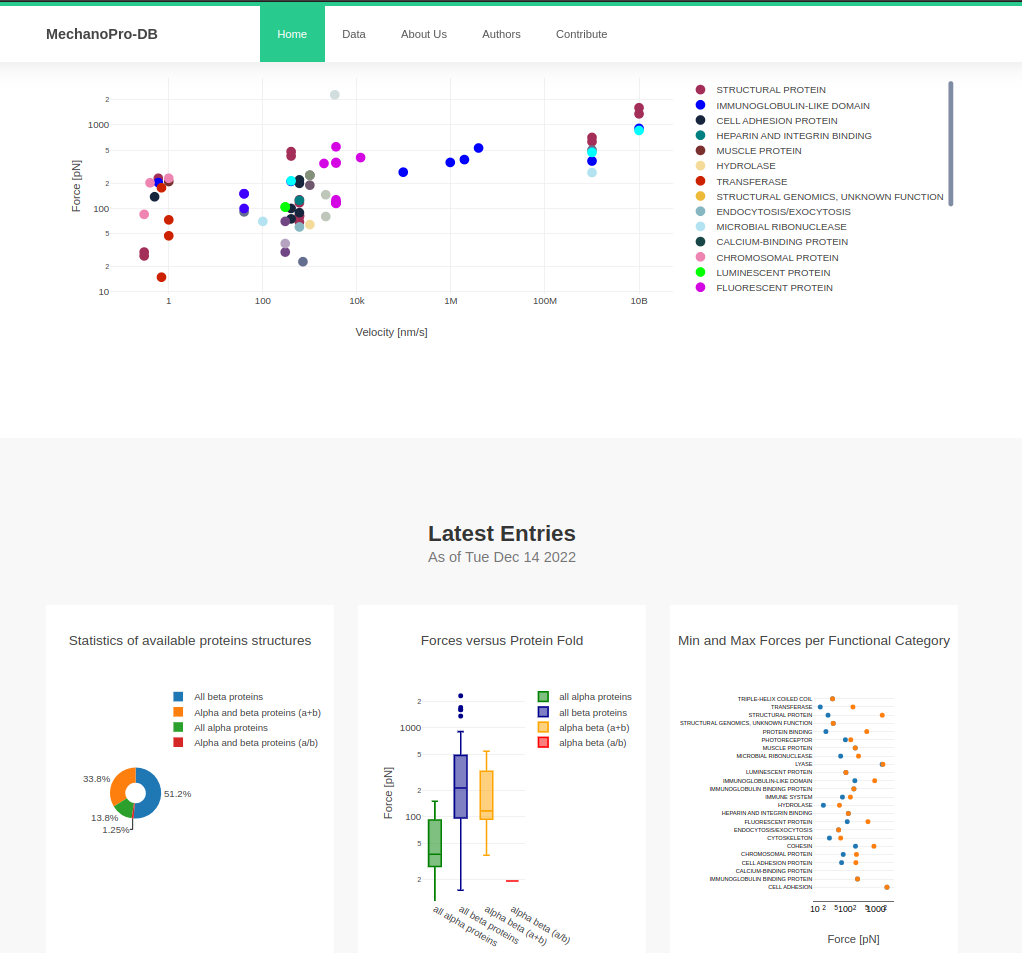
About MechanoPro-DataBase
Mechanical forces are involved in a variety of biological processes, including protein degradation, cell adhesion, tissue organization,
and muscle contraction in multicellular organisms.
Proteins are the main participants in these events.
These nanomachines are subjected of exert mechanical force in diverse biological processes at cellular, subcellular and even molecular level.
As a result, studying the mechanical responses of these biomolecules will yield a wealth of knowledge about their structure and function.
The use of nanotechnologies, such as optical tweezers and atomic force microscopy, at the single molecule level to understand biological phenomena has
accelerated experimental characterization of mechanical reaction of biomolecules and have made significant strides in understanding. Steered Molecular Dynamics simulations,
on the other hand enable us to access atomistic details.
MechanoProDataBase stores experimental data from force spectrsocpy experiments and computational data from simulatons.
You will find below a detailed tutorial that will help you get familiar with MechanoProDB.
A the bottom of the page, a video shows you the different steps that have been detailed.

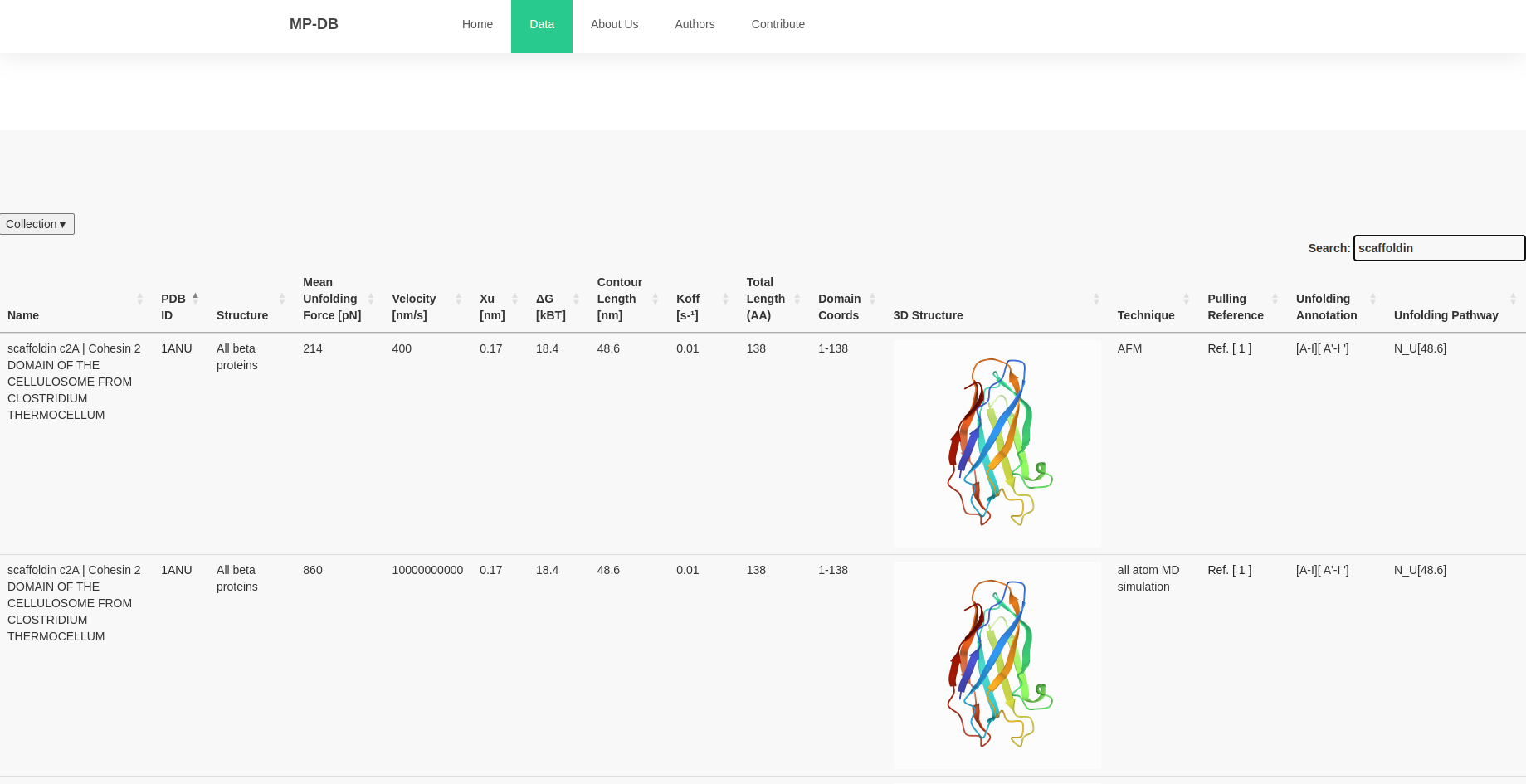
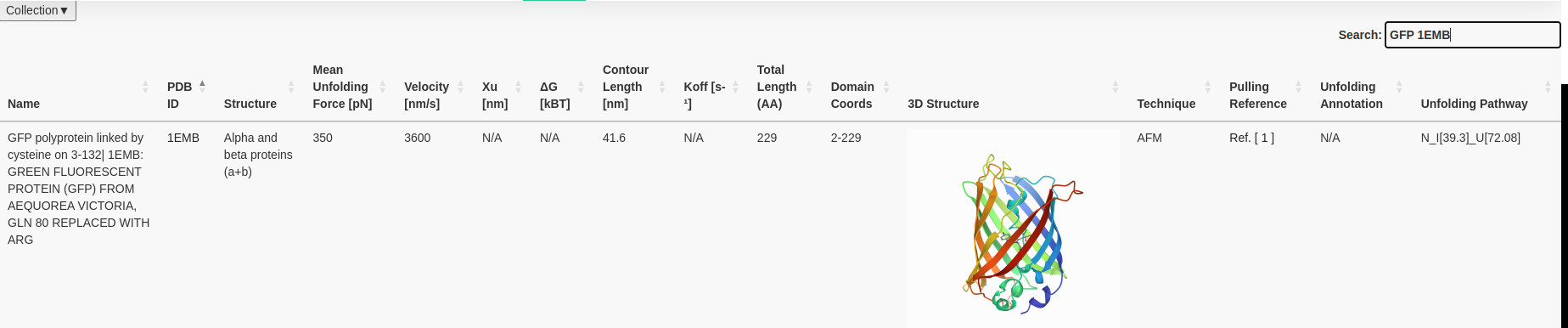 You have the possibility to display the information you want to see by clicking at collections (top left) and selecting or unselection the columns you do
not want to see.
This tab, gives you also the possibility to download the available data to CSV.
NB: Since one AFM based force spectroscopy experiment can result in different unfolding forces at different velocities/loading rates, we display all available data points provided within the manuscript.
You have the possibility to display the information you want to see by clicking at collections (top left) and selecting or unselection the columns you do
not want to see.
This tab, gives you also the possibility to download the available data to CSV.
NB: Since one AFM based force spectroscopy experiment can result in different unfolding forces at different velocities/loading rates, we display all available data points provided within the manuscript.